checkpointing: Stan
chkpt_stan.RmdThe following examples walk through using chkptstanr with the Stan
The basic idea is to (1) write a custom Stan model
(done by the user), (2) fit the model with cmdstanr (with
the desired number of checkpoints), and then (3) return a
cmststanr object. All but step (1) is done internally, so
the workflow is very similar to using cmdstanr.
Example 1: Eight Schools
Storage
The initial overhead is to create a folder that will store the checkpoints, i.e.,
path <- create_folder(folder_name = "chkpt_folder_m1")Stan Model
Next is the Stan model:
stan_code <- "
data {
int<lower=0> n;
real y[n];
real<lower=0> sigma[n];
}
parameters {
real mu;
real<lower=0> tau;
vector[n] eta;
}
transformed parameters {
vector[n] theta;
theta = mu + tau * eta;
}
model {
target += normal_lpdf(eta | 0, 1);
target += normal_lpdf(y | theta, sigma);
}
"Stan Data
When using chkpt_stan(), this requires supplying a list
to the data argument, much like using rstan.
Model Fitting
2 Checkpoints
To show the basic idea of checkpointing, the following was stopped after 2 checkpoints.
fit_m1 <- chkpt_stan(model_code = stan_code,
data = stan_data,
iter_warmup = 1000,
iter_sampling = 1000,
iter_per_chkpt = 250,
path = path)
#> Compiling Stan program...
#> Initial Warmup (Typical Set)
#> Chkpt: 1 / 8; Iteration: 250 / 2000 (warmup)
#> Chkpt: 2 / 8; Iteration: 500 / 2000 (warmup)Finish Sampling
To finish the remaining 6 checkpoints run the same code, i.e.,
fit_m1 <- chkpt_stan(model_code = stan_code,
data = stan_data,
iter_warmup = 1000,
iter_sampling = 1000,
iter_per_chkpt = 250,
path = path)
#> Sampling next checkpoint
#> Chkpt: 3 / 8; Iteration: 750 / 2000 (warmup)
#> Chkpt: 4 / 8; Iteration: 1000 / 2000 (warmup)
#> Chkpt: 5 / 8; Iteration: 1250 / 2000 (sample)
#> Chkpt: 6 / 8; Iteration: 1500 / 2000 (sample)
#> Chkpt: 7 / 8; Iteration: 1750 / 2000 (sample)
#> Chkpt: 8 / 8; Iteration: 2000 / 2000 (sample)
#> Checkpointing completeCombine Draws
Each checkpoint contains 250 draws from the posterior. These need to
be combined with combine_chkpt_draws(), i.e.,
draws <- combine_chkpt_draws(fit_m1)We developed chkptstanr to work seamlessly with the
Stan ecosystem. The object draws has been
constructed to mimic what is provided when using
cmdstanr directly.
combine_chkpt_draws(fit_m1)
#> # A draws_array: 1000 iterations, 2 chains, and 19 variables
#> , , variable = lp__
#>
#> chain
#> iteration 1 2
#> 1 -34 -43
#> 2 -37 -41
#> 3 -36 -39
#> 4 -38 -38
#> 5 -38 -41
#>
#> , , variable = mu
#>
#> chain
#> iteration 1 2
#> 1 5.2 2.6
#> 2 11.3 6.7
#> 3 -2.7 5.3
#> 4 -2.9 3.7
#> 5 -2.7 14.2
#>
#> , , variable = tau
#>
#> chain
#> iteration 1 2
#> 1 23.3 2.61
#> 2 6.7 0.21
#> 3 12.7 4.44
#> 4 21.1 7.29
#> 5 18.8 10.94
#>
#> , , variable = eta[1]
#>
#> chain
#> iteration 1 2
#> 1 0.10 -0.61
#> 2 0.89 -0.87
#> 3 1.62 0.83
#> 4 1.99 0.84
#> 5 -0.16 1.22
#>
#> # ... with 995 more iterations, and 15 more variablesSummary
draws can then be used with the R package
posterior
posterior::summarise_draws(draws)
#> # A tibble: 19 x 10
#> variable mean median sd mad q5 q95 rhat ess_bulk ess_tail
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 lp__ -39.5 -39.2 2.59 2.58 -44.2 -35.9 1.00 640. 1008.
#> 2 mu 7.77 7.92 5.48 5.10 -1.43 16.0 1.01 530. 325.
#> 3 tau 6.82 5.32 5.75 4.71 0.434 18.7 1.00 649. 658.
#> 4 eta[1] 0.383 0.413 0.929 0.909 -1.20 1.87 1.00 1650. 1233.
#> 5 eta[2] -0.00335 -0.00816 0.841 0.814 -1.34 1.40 1.00 1443. 1307.
#> 6 eta[3] -0.176 -0.174 0.931 0.906 -1.67 1.42 1.00 1829. 1424.
#> 7 eta[4] -0.00521 0.000856 0.862 0.841 -1.47 1.39 1.00 1565. 1407.
#> 8 eta[5] -0.312 -0.350 0.873 0.835 -1.72 1.24 1.00 1661. 1616.
#> 9 eta[6] -0.193 -0.190 0.889 0.909 -1.59 1.28 1.00 1915. 1404.
#> 10 eta[7] 0.387 0.358 0.876 0.864 -1.09 1.81 1.00 1574. 1370.
#> 11 eta[8] 0.0805 0.0611 0.970 0.960 -1.51 1.66 1.00 1031. 1236.
#> 12 theta[1] 11.5 10.2 8.29 6.99 0.268 26.4 1.00 1042. 728.
#> 13 theta[2] 7.87 7.87 6.20 5.66 -2.27 17.8 1.00 1549. 1515.
#> 14 theta[3] 6.01 6.63 8.25 6.63 -8.69 18.1 1.00 1102. 1075.
#> 15 theta[4] 7.75 7.76 6.65 5.96 -3.06 18.9 1.00 1674. 1210.
#> 16 theta[5] 5.05 5.70 6.44 5.75 -7.06 14.4 1.00 1405. 1416.
#> 17 theta[6] 6.21 6.60 6.92 6.15 -5.98 16.9 1.00 1890. 1195.
#> 18 theta[7] 10.8 10.1 6.71 6.03 0.992 23.1 1.00 1497. 1767.
#> 19 theta[8] 8.35 8.41 7.72 6.66 -3.88 20.7 1.00 1081. 1075.Visualization with bayesplot
The popular R package bayesplot can
also be used.
bayesplot::mcmc_trace(draws) +
geom_vline(xintercept = seq(0, 1000, 250),
alpha = 0.25,
size = 2)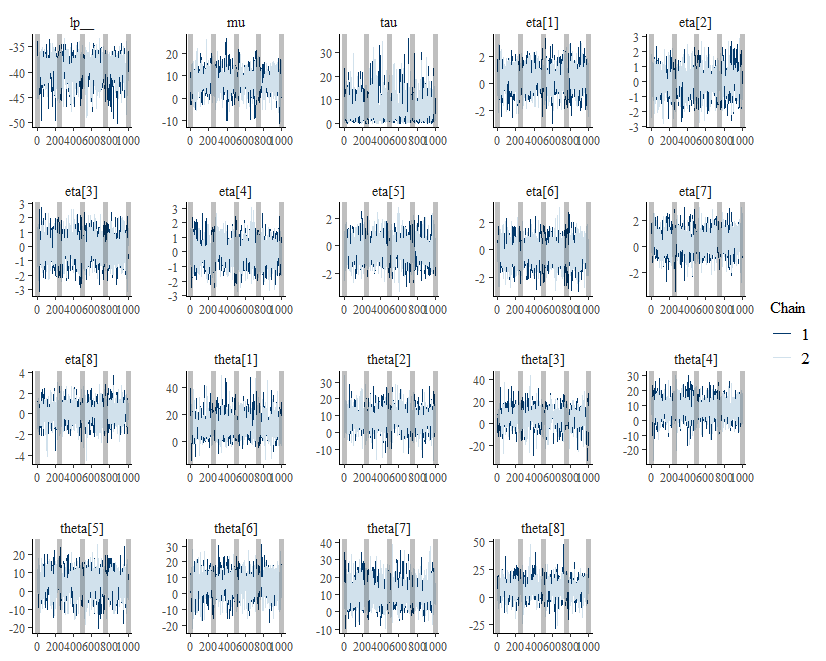
This vertical lines are placed at each checkpoint.