Impute values, assuming a multivariate normal distribution, with the posterior predictive distribution. For binary, ordinal, and mixed (a combination of discrete and continuous) data, the values are first imputed for the latent data and then converted to the original scale.
mvn_imputation( Y, type = "continuous", iter = 1000, progress = TRUE, save_all = FALSE )
Arguments
| Y | Matrix (or data frame) of dimensions n (observations) by p (variables). |
|---|---|
| type | Character string. Which type of data for |
| iter | Number of iterations (posterior samples; defaults to 1000). |
| progress | Logical. Should a progress bar be included (defaults to |
| save_all | Logical. Should each imputed dataset be stored
(defaults to |
Value
An object of class mvn_imputation:
YThe last imputed dataset.ppd_missingA matrix of dimensionsiterby the number of missing values.ppd_meanA vector including the means of the posterior predictive distribution for the missing values.Y_allAn 3D array withitermatrices of dimensions n by p (NULLwhensave_all = FALSE).
Details
Missing values are imputed with the approach described in Hoff (2009)
.
The basic idea is to impute the missing values with the respective posterior pedictive distribution,
given the observed data, as the model is being estimated. Note that the default is TRUE,
but this ignored when there are no missing values. If set to FALSE, and there are missing
values, list-wise deletion is performed with na.omit.
References
Hoff PD (2009). A first course in Bayesian statistical methods, volume 580. Springer.
Examples
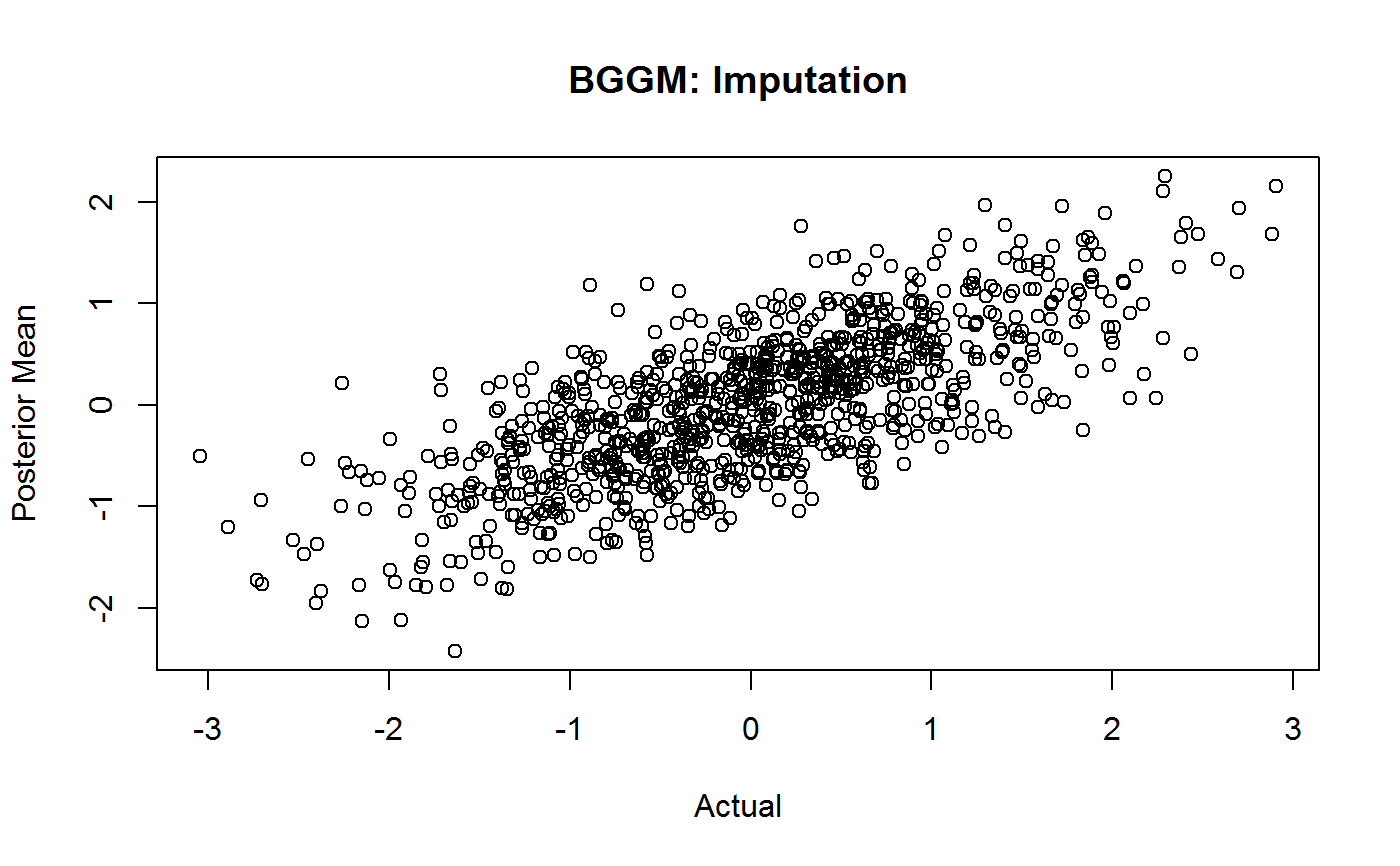
# \donttest{ # obs n <- 5000 # n missing n_missing <- 1000 # variables p <- 16 # data Y <- MASS::mvrnorm(n, rep(0, p), ptsd_cor1) # for checking Ymain <- Y # all possible indices indices <- which(matrix(0, n, p) == 0, arr.ind = TRUE) # random sample of 1000 missing values na_indices <- indices[sample(5:nrow(indices), size = n_missing, replace = FALSE),] # fill with NA Y[na_indices] <- NA # missing = 1 Y_miss <- ifelse(is.na(Y), 1, 0) # true values (to check) true <- unlist(sapply(1:p, function(x) Ymain[which(Y_miss[,x] == 1),x] )) # impute fit_missing <- mvn_imputation(Y, progress = FALSE, iter = 250) print(fit_missing, n_rows = 20)#> BGGM: Bayesian Gaussian Graphical Models #> --- #> Multivariate Normal Imputation #> --- #> Estimates: #> #> Value Post.mean Post.sd Cred.lb Cred.ub #> 57--1 1.937 0.493 0.849 2.878 #> 244--1 -0.401 0.501 -1.390 0.581 #> 273--1 -0.437 0.502 -1.388 0.538 #> 317--1 -0.014 0.515 -0.983 0.965 #> 403--1 0.939 0.509 -0.002 1.913 #> 449--1 -1.502 0.507 -2.377 -0.455 #> 461--1 -0.144 0.525 -1.146 0.914 #> 556--1 0.709 0.508 -0.293 1.591 #> 558--1 -0.525 0.513 -1.647 0.470 #> 686--1 -2.127 0.489 -3.044 -1.172 #> 777--1 0.707 0.514 -0.298 1.655 #> 824--1 -0.273 0.480 -1.191 0.565 #> 872--1 -0.707 0.557 -1.806 0.354 #> 908--1 -1.073 0.550 -2.259 -0.067 #> 948--1 -0.187 0.510 -1.107 0.871 #> 958--1 0.448 0.539 -0.546 1.492 #> 1026--1 -1.336 0.576 -2.443 -0.211 #> 1074--1 0.063 0.542 -1.019 1.062 #> 1116--1 0.222 0.536 -0.757 1.265 #> 1139--1 -1.627 0.483 -2.570 -0.668# plot plot(x = true, y = fit_missing$ppd_mean, main = "BGGM: Imputation", xlab = "Actual", ylab = "Posterior Mean")# }